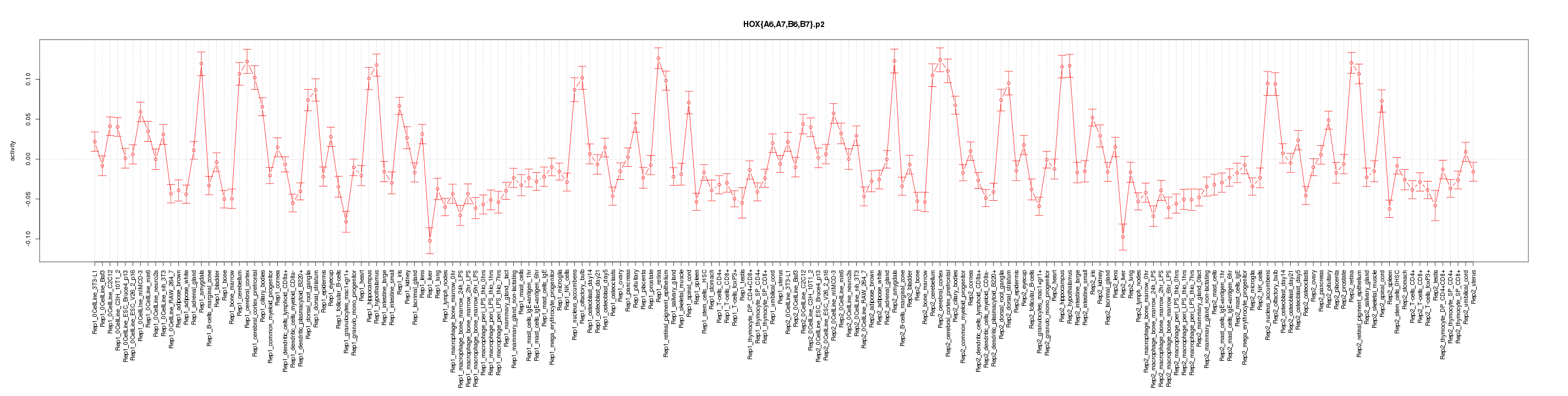
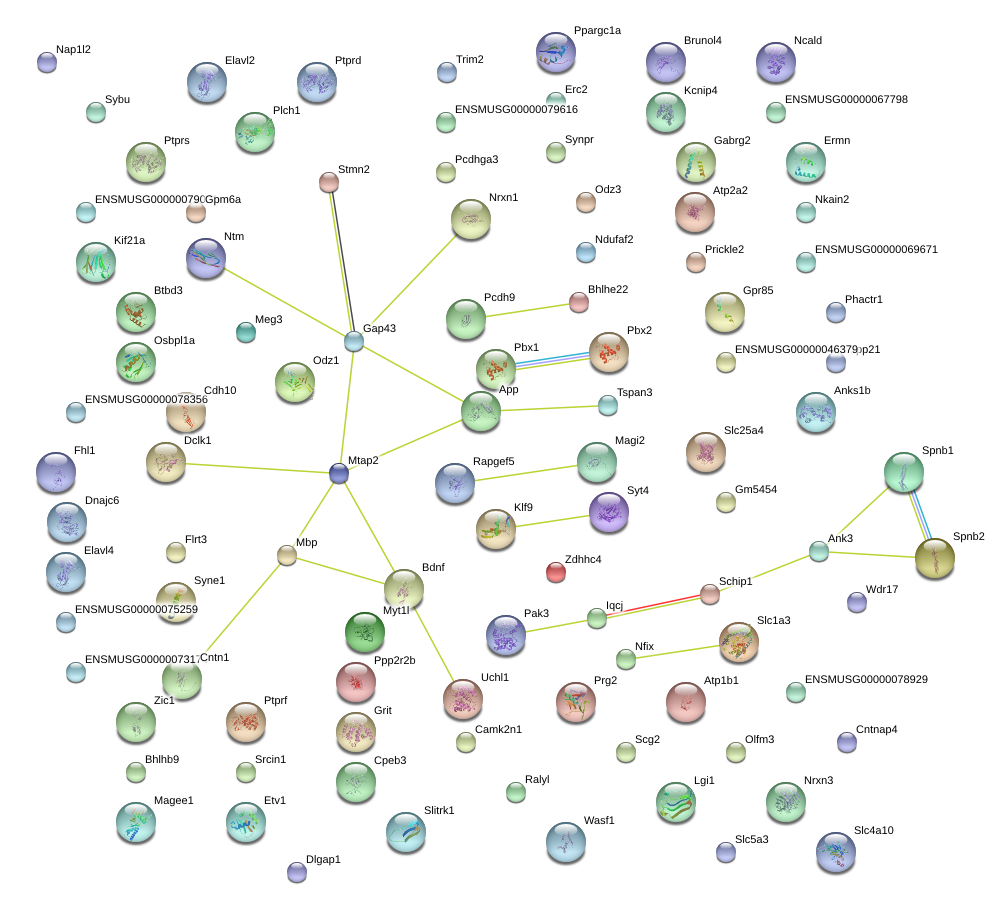
|
chr8_+_56040211
|
224.941
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr8_+_56040445
|
216.821
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr8_+_56040553
|
199.388
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr8_+_56040080
|
187.589
|
NM_153581
|
Gpm6a
|
glycoprotein m6a
|
|
chr8_+_56040193
|
183.387
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr13_+_42776161
|
131.633
|
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr3_+_68376319
|
111.527
|
|
Schip1
|
schwannomin interacting protein 1
|
|
chr10_+_69487700
|
110.800
|
|
Ank3
|
ankyrin 3, epithelial
|
|
chr17_-_90487479
|
109.531
|
|
Nrxn1
|
neurexin I
|
|
chr16_+_7069971
|
108.774
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr11_-_30000031
|
101.099
|
|
Spnb2
|
spectrin beta 2
|
|
chr3_+_67696141
|
100.219
|
NM_001113419
NM_177585
|
Iqcj-schip1
Iqcj
|
IQ motif containing J-schwannomin interacting protein 1 read-through transcript
IQ motif containing J
|
|
chr3_+_17695743
|
99.031
|
|
6430547I21Rik
|
RIKEN cDNA 6430547I21 gene
|
|
chrX_+_10173557
|
98.994
|
|
|
|
|
chr13_+_42775991
|
98.417
|
NM_198419
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr9_-_91255831
|
95.085
|
|
Zic1
|
zinc finger protein of the cerebellum 1
|
|
chrX_+_54032914
|
93.826
|
|
Fhl1
|
four and a half LIM domains 1
|
|
chrX_+_132424640
|
93.624
|
|
Bhlhb9
|
basic helix-loop-helix domain containing, class B9
|
|
chr14_+_28689289
|
93.342
|
|
Erc2
|
ELKS/RAB6-interacting/CAST family member 2
|
|
chr16_+_7069927
|
92.902
|
NM_183188
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr14_-_94287950
|
90.768
|
NM_001081377
|
Pcdh9
|
protocadherin 9
|
|
chrX_+_54032858
|
89.546
|
NM_001077362
|
Fhl1
|
four and a half LIM domains 1
|
|
chrX_-_100381973
|
84.547
|
NM_008671
|
Nap1l2
|
nucleosome assembly protein 1-like 2
|
|
chr3_+_68376289
|
84.224
|
NM_013928
|
Schip1
|
schwannomin interacting protein 1
|
|
chr12_+_30219769
|
83.152
|
NM_001093776
|
Myt1l
|
myelin transcription factor 1-like
|
|
chrX_-_100381703
|
82.399
|
|
Nap1l2
|
nucleosome assembly protein 1-like 2
|
|
chr10_+_40657980
|
81.523
|
|
Wasf1
|
WASP family 1
|
|
chr15_+_91991704
|
79.535
|
NM_001159647
|
Cntn1
|
contactin 1
|
|
chr5_-_122907002
|
78.475
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr15_-_90764873
|
77.834
|
|
Kif21a
|
kinesin family member 21A
|
|
chr4_+_101180625
|
76.079
|
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr3_+_13946371
|
75.925
|
NM_001163329
NM_001163330
|
Ralyl
|
RALY RNA binding protein-like
|
|
chr10_-_31410391
|
74.916
|
|
Nkain2
|
Na+/K+ transporting ATPase interacting 2
|
|
chr10_+_90038471
|
73.930
|
NM_001177397
|
Anks1b
|
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr18_-_25637786
|
72.173
|
|
Celf4
|
CUGBP, Elav-like family member 4
|
|
chr5_-_122906470
|
70.216
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr16_-_42340613
|
69.944
|
NM_008083
|
Gap43
|
growth associated protein 43
|
|
chr1_-_79436564
|
69.934
|
NM_009129
|
Scg2
|
secretogranin II
|
|
chr11_-_41814008
|
69.550
|
NM_008073
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2
|
|
chr18_+_82754829
|
69.413
|
|
Mbp
|
myelin basic protein
|
|
chr9_-_112090150
|
69.136
|
NM_033264
NM_001177615
NM_001177620
NM_028755
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21
|
|
chr4_+_138012860
|
68.754
|
|
Camk2n1
|
calcium/calmodulin-dependent protein kinase II inhibitor 1
|
|
chr8_+_56122680
|
68.584
|
|
Gpm6a
|
glycoprotein m6a
|
|
chr6_-_13785233
|
67.377
|
|
Gpr85
|
G protein-coupled receptor 85
|
|
chr14_-_109313298
|
67.326
|
|
Slitrk1
|
SLIT and NTRK-like family, member 1
|
|
chr18_-_42900793
|
66.988
|
|
Ppp2r2b
|
protein phosphatase 2 (formerly 2A), regulatory subunit B (PR 52), beta isoform
|
|
chr18_+_82754586
|
66.914
|
|
Mbp
|
myelin basic protein
|
|
chr18_-_25637339
|
66.843
|
|
Celf4
|
CUGBP, Elav-like family member 4
|
|
chr18_-_12914177
|
66.785
|
|
Osbpl1a
|
oxysterol binding protein-like 1A
|
|
chr12_+_110782654
|
66.178
|
|
Meg3
|
maternally expressed 3
|
|
chr14_-_109313333
|
66.136
|
|
Slitrk1
|
SLIT and NTRK-like family, member 1
|
|
chr15_+_18750083
|
65.941
|
NM_009865
|
Cdh10
|
cadherin 10
|
|
chr2_-_140485267
|
65.631
|
|
Flrt3
|
fibronectin leucine rich transmembrane protein 3
|
|
chr16_-_84955086
|
65.225
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr9_-_29770699
|
64.138
|
NM_172290
|
Ntm
|
neurotrimin
|
|
chr18_+_82754377
|
63.367
|
|
Mbp
|
myelin basic protein
|
|
chr11_-_41813792
|
62.914
|
NM_177408
|
Gabrg2
|
gamma-aminobutyric acid (GABA) A receptor, subunit gamma 2
|
|
chr1_-_170085342
|
61.043
|
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr19_+_23241546
|
60.918
|
|
Klf9
|
Kruppel-like factor 9
|
|
chr6_-_13789847
|
57.884
|
NM_145066
|
Gpr85
|
G protein-coupled receptor 85
|
|
chr5_+_19413335
|
55.774
|
NM_001170745
NM_015823
|
Magi2
|
membrane associated guanylate kinase, WW and PDZ domain containing 2
|
|
chr5_-_49280703
|
55.494
|
NM_030265
|
Kcnip4
|
Kv channel interacting protein 4
|
|
chr1_+_66368571
|
55.194
|
|
Mtap2
|
microtubule-associated protein 2
|
|
chr3_+_17954324
|
55.179
|
NM_021560
|
Bhlhe22
|
basic helix-loop-helix family, member e22
|
|
chr8_-_55804030
|
54.413
|
NM_028220
|
Wdr17
|
WD repeat domain 17
|
|
chr9_-_55983967
|
54.122
|
|
Tspan3
|
tetraspanin 3
|
|
chr4_-_109960078
|
53.727
|
NM_001038698
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr3_+_8509479
|
52.999
|
NM_025285
|
Stmn2
|
stathmin-like 2
|
|
chr1_-_166368439
|
52.287
|
|
Atp1b1
|
ATPase, Na+/K+ transporting, beta 1 polypeptide
|
|
chr14_+_14286471
|
51.612
|
NM_001163032
|
Synpr
|
synaptoporin
|
|
chr3_-_83967952
|
51.612
|
|
Trim2
|
tripartite motif-containing 2
|
|
chr5_-_51848420
|
51.347
|
|
Ppargc1a
|
peroxisome proliferative activated receptor, gamma, coactivator 1 alpha
|
|
chr3_-_63654912
|
51.239
|
NM_001177732
NM_001177733
NM_183191
|
Plch1
|
phospholipase C, eta 1
|
|
chr15_-_8660729
|
50.205
|
NM_148938
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr15_-_37389073
|
49.865
|
NM_001170867
NM_001170868
|
Ncald
|
neurocalcin delta
|
|
chrX_-_40627962
|
49.733
|
NM_011855
|
Odz1
|
odd Oz/ten-m homolog 1 (Drosophila)
|
|
chr2_-_57905140
|
49.514
|
NM_029972
|
Ermn
|
ermin, ERM-like protein
|
|
chr15_-_44584003
|
49.244
|
NM_178765
|
Sybu
|
syntabulin (syntaxin-interacting)
|
|
chr11_-_97372344
|
49.095
|
|
Srcin1
|
SRC kinase signaling inhibitor 1
|
|
chr8_-_47292973
|
49.073
|
|
Slc25a4
|
solute carrier family 25 (mitochondrial carrier, adenine nucleotide translocator), member 4
|
|
chr3_+_114783882
|
47.505
|
NM_153157
|
Olfm3
|
olfactomedin 3
|
|
chr12_+_118997623
|
47.424
|
|
Rapgef5
|
Rap guanine nucleotide exchange factor (GEF) 5
|
|
chr19_+_38339167
|
47.292
|
NM_020278
|
Lgi1
|
leucine-rich repeat LGI family, member 1
|
|
chr3_+_55265654
|
47.085
|
NM_001111051
NM_001111052
NM_001195539
NM_001195540
|
Dclk1
|
doublecortin-like kinase 1
|
|
chr5_+_67067359
|
47.073
|
NM_011670
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1
|
|
chr16_+_7409879
|
46.789
|
|
Rbfox1
|
RNA binding protein, fox-1 homolog (C. elegans) 1
|
|
chr9_+_31923720
|
46.289
|
NM_001195632
|
Arhgap32
|
Rho GTPase activating protein 32
|
|
chr16_+_92086695
|
45.808
|
|
Slc5a3
|
solute carrier family 5 (inositol transporters), member 3
|
|
chr17_+_70871444
|
45.743
|
NM_001128180
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr18_-_31606908
|
45.614
|
NM_009308
|
Syt4
|
synaptotagmin IV
|
|
chrX_+_102318792
|
45.508
|
|
Magee1
|
melanoma antigen, family E, 1
|
|
chr10_+_4978808
|
45.118
|
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr2_+_109532592
|
44.907
|
NM_001048141
|
Bdnf
|
brain derived neurotrophic factor
|
|
chr6_-_92431385
|
44.892
|
NM_001134460
|
Prickle2
|
prickle homolog 2 (Drosophila)
|
|
chr12_+_110834872
|
44.876
|
|
1110006E14Rik
|
RIKEN cDNA 1110006E14 gene
|
|
chr8_+_115093942
|
44.675
|
NM_130457
|
Cntnap4
|
contactin associated protein-like 4
|
|
chr18_+_37925619
|
44.507
|
|
Pcdhga12
|
protocadherin gamma subfamily A, 12
|
|
chr2_+_138104219
|
44.489
|
NM_145534
|
Btbd3
|
BTB (POZ) domain containing 3
|
|
chr19_-_37250472
|
44.480
|
|
Cpeb3
|
cytoplasmic polyadenylation element binding protein 3
|
|
chr12_+_110800212
|
44.472
|
|
Meg3
|
maternally expressed 3
|
|
chr12_+_91570569
|
44.273
|
|
Nrxn3
|
neurexin III
|
|
chrX_+_139997806
|
44.053
|
NM_008778
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr4_-_76240202
|
43.897
|
NM_011211
|
Ptprd
|
protein tyrosine phosphatase, receptor type, D
|
|
chr2_+_61884596
|
43.868
|
NM_033552
|
Slc4a10
|
solute carrier family 4, sodium bicarbonate cotransporter-like, member 10
|
|
chr12_+_30213224
|
43.364
|
NM_001093775
NM_001093778
NM_008666
|
Myt1l
|
myelin transcription factor 1-like
|
|
chr8_-_87297283
|
42.928
|
|
Nfix
|
nuclear factor I/X
|
|
chr4_-_91066674
|
42.600
|
NM_207685
|
Elavl2
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 2 (Hu antigen B)
|
|
chr12_+_39510297
|
42.545
|
NM_001163154
|
Etv1
|
ets variant gene 1
|
|
chr15_+_81626455
|
42.536
|
|
Zc3h7b
|
zinc finger CCCH type containing 7B
|
|
chr8_-_47292863
|
42.465
|
|
|
|
|
chr7_+_107422781
|
42.369
|
|
Pgm2l1
|
phosphoglucomutase 2-like 1
|
|
chr17_+_70871393
|
42.327
|
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr16_+_43247396
|
42.295
|
NM_019778
NM_181058
|
Zbtb20
|
zinc finger and BTB domain containing 20
|
|
chr16_-_84955488
|
42.156
|
|
App
|
amyloid beta (A4) precursor protein
|
|
chr3_+_55265811
|
42.113
|
|
Dclk1
|
doublecortin-like kinase 1
|
|
chrX_+_155897136
|
41.985
|
|
Mtap7d2
|
MAP7 domain containing 2
|
|
chr15_-_8660479
|
41.494
|
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr10_+_98486068
|
41.173
|
|
Atp2b1
|
ATPase, Ca++ transporting, plasma membrane 1
|
|
chr14_+_13073221
|
41.092
|
|
Ptprg
|
protein tyrosine phosphatase, receptor type, G
|
|
chr13_+_59234737
|
40.272
|
|
Ntrk2
|
neurotrophic tyrosine kinase, receptor, type 2
|
|
chr4_-_109959185
|
40.114
|
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr10_-_116782524
|
40.051
|
|
Cpsf6
|
cleavage and polyadenylation specific factor 6
|
|
chr17_-_85461840
|
40.038
|
|
Prepl
|
prolyl endopeptidase-like
|
|
chr3_-_122241909
|
39.970
|
|
Fnbp1l
|
formin binding protein 1-like
|
|
chr3_+_136598184
|
39.949
|
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform
|
|
chr10_+_69488049
|
39.798
|
|
Ank3
|
ankyrin 3, epithelial
|
|
chr11_-_73137838
|
39.775
|
|
Aspa
|
aspartoacylase
|
|
chr8_-_67071497
|
39.730
|
|
|
|
|
chr3_+_136598485
|
39.349
|
|
Ppp3ca
|
protein phosphatase 3, catalytic subunit, alpha isoform
|
|
chr9_-_83003967
|
39.212
|
|
|
|
|
chr5_-_122907269
|
39.021
|
|
Atp2a2
|
ATPase, Ca++ transporting, cardiac muscle, slow twitch 2
|
|
chr2_+_59192308
|
38.951
|
|
Pkp4
|
plakophilin 4
|
|
chr1_-_42749940
|
38.885
|
|
2610017I09Rik
|
RIKEN cDNA 2610017I09 gene
|
|
chr18_+_37834003
|
38.825
|
|
Pcdhga3
|
protocadherin gamma subfamily A, 3
|
|
chr11_-_79317471
|
38.586
|
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr14_-_109313447
|
38.554
|
NM_199065
|
Slitrk1
|
SLIT and NTRK-like family, member 1
|
|
chr11_-_79317583
|
38.026
|
NM_019409
|
Omg
|
oligodendrocyte myelin glycoprotein
|
|
chr9_+_75159201
|
38.025
|
NM_010313
|
Gnb5
|
guanine nucleotide binding protein (G protein), beta 5
|
|
chr3_-_126701367
|
37.738
|
NM_178655
|
Ank2
|
ankyrin 2, brain
|
|
chr9_-_112134930
|
37.506
|
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21
|
|
chr12_+_110783786
|
37.390
|
|
Meg3
|
maternally expressed 3
|
|
chr13_+_42961598
|
37.377
|
|
Phactr1
|
phosphatase and actin regulator 1
|
|
chr10_+_4978047
|
37.134
|
|
Syne1
|
synaptic nuclear envelope 1
|
|
chr9_-_112134984
|
36.982
|
NM_001177616
|
Arpp21
|
cyclic AMP-regulated phosphoprotein, 21
|
|
chr15_-_8660462
|
36.980
|
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr11_-_69648350
|
36.899
|
NM_198862
|
Nlgn2
|
neuroligin 2
|
|
chr2_+_97307652
|
36.799
|
NM_178725
|
Lrrc4c
|
leucine rich repeat containing 4C
|
|
chr11_+_117623703
|
36.748
|
|
Tnrc6c
|
trinucleotide repeat containing 6C
|
|
chr15_-_8660442
|
36.743
|
|
Slc1a3
|
solute carrier family 1 (glial high affinity glutamate transporter), member 3
|
|
chr9_+_57810230
|
36.635
|
|
Sema7a
|
sema domain, immunoglobulin domain (Ig), and GPI membrane anchor, (semaphorin) 7A
|
|
chr19_+_10525829
|
36.561
|
|
|
|
|
chr17_+_71168939
|
36.144
|
|
Dlgap1
|
discs, large (Drosophila) homolog-associated protein 1
|
|
chr2_-_77357674
|
35.566
|
NM_001113400
|
Zfp385b
|
zinc finger protein 385B
|
|
chr18_+_57628139
|
35.280
|
NM_001134697
|
Ctxn3
|
cortexin 3
|
|
chr7_+_107789414
|
35.051
|
|
Rab6
|
RAB6, member RAS oncogene family
|
|
chr1_-_134603848
|
34.976
|
NM_001160318
|
Nfasc
|
neurofascin
|
|
chr18_+_38476622
|
34.631
|
|
|
|
|
chr18_+_82753560
|
34.524
|
|
Mbp
|
myelin basic protein
|
|
chr10_-_63552994
|
34.335
|
NM_178678
|
Lrrtm3
|
leucine rich repeat transmembrane neuronal 3
|
|
chrX_+_162782484
|
34.081
|
NM_001177955
NM_001177956
NM_001177957
NM_001177958
NM_001177959
NM_001177960
|
Gpm6b
|
glycoprotein m6b
|
|
chr2_+_174171943
|
34.077
|
|
|
|
|
chr11_+_87032504
|
33.998
|
|
Trim37
|
tripartite motif-containing 37
|
|
chr14_+_120830603
|
33.876
|
|
Mbnl2
|
muscleblind-like 2
|
|
chr4_+_101180580
|
33.648
|
NM_001164584
NM_001164585
|
Dnajc6
|
DnaJ (Hsp40) homolog, subfamily C, member 6
|
|
chr1_+_34364633
|
33.452
|
|
Dst
|
dystonin
|
|
chr2_-_79969095
|
33.383
|
NM_016744
|
Pde1a
|
phosphodiesterase 1A, calmodulin-dependent
|
|
chr17_-_90436488
|
33.264
|
|
Nrxn1
|
neurexin I
|
|
chr17_+_17539832
|
33.248
|
|
Lix1
|
limb expression 1 homolog (chicken)
|
|
chr12_-_32398730
|
33.163
|
NM_175191
|
Gpr22
|
G protein-coupled receptor 22
|
|
chr5_+_67067497
|
33.128
|
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1
|
|
chr8_-_70342105
|
32.949
|
NM_027626
|
Psd3
|
pleckstrin and Sec7 domain containing 3
|
|
chr3_+_55586431
|
32.728
|
NM_010750
|
Mab21l1
|
mab-21-like 1 (C. elegans)
|
|
chr5_+_67067477
|
32.490
|
|
Uchl1
|
ubiquitin carboxy-terminal hydrolase L1
|
|
chr18_+_37821598
|
32.301
|
NM_033584
|
Pcdhga1
|
protocadherin gamma subfamily A, 1
|
|
chr17_+_17539908
|
32.140
|
|
Lix1
|
limb expression 1 homolog (chicken)
|
|
chr1_-_170085798
|
32.112
|
|
Pbx1
|
pre B-cell leukemia transcription factor 1
|
|
chr4_-_86873866
|
31.884
|
NM_001110240
|
Slc24a2
|
solute carrier family 24 (sodium/potassium/calcium exchanger), member 2
|
|
chr19_-_7500486
|
31.855
|
|
|
|
|
chr18_+_23573939
|
31.819
|
|
Dtna
|
dystrobrevin alpha
|
|
chr7_+_134094045
|
31.528
|
NM_144926
|
Sez6l2
|
seizure related 6 homolog like 2
|
|
chr14_+_55506625
|
31.495
|
|
Bcl2l2
|
BCL2-like 2
|
|
chr1_-_194677940
|
31.435
|
NM_001177794
|
Sertad4
|
SERTA domain containing 4
|
|
chr12_-_42212538
|
31.327
|
|
Lrrn3
|
leucine rich repeat protein 3, neuronal
|
|
chrM_+_11741
|
31.029
|
|
ND5
|
NADH dehydrogenase subunit 5
|
|
chr2_-_77541328
|
30.985
|
NM_001113399
|
Zfp385b
|
zinc finger protein 385B
|
|
chr5_-_23012407
|
30.752
|
|
Srpk2
|
serine/arginine-rich protein specific kinase 2
|
|
chrX_+_140100287
|
30.689
|
NM_001195049
|
Pak3
|
p21 protein (Cdc42/Rac)-activated kinase 3
|
|
chr2_-_115890747
|
30.681
|
NM_001136072
NM_001159567
NM_001159568
NM_010825
|
Meis2
|
Meis homeobox 2
|
|
chrX_+_38754568
|
30.590
|
|
Gria3
|
glutamate receptor, ionotropic, AMPA3 (alpha 3)
|
|
chr11_+_52202137
|
30.291
|
|
Vdac1
|
voltage-dependent anion channel 1
|
|
chr11_-_41996141
|
30.127
|
NM_010250
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1
|
|
chrX_-_22797942
|
30.049
|
|
Klhl13
|
kelch-like 13 (Drosophila)
|
|
chr8_-_37631532
|
29.767
|
|
Dlc1
|
deleted in liver cancer 1
|
|
chr3_-_146432922
|
29.673
|
|
Prkacb
|
protein kinase, cAMP dependent, catalytic, beta
|
|
chr11_+_42234760
|
29.647
|
|
Gabrb2
|
gamma-aminobutyric acid (GABA) A receptor, subunit beta 2
|
|
chr10_+_90039435
|
29.586
|
NM_001177396
NM_001177398
NM_181398
|
Ipw
Anks1b
|
imprinted gene in the Prader-Willi syndrome region
ankyrin repeat and sterile alpha motif domain containing 1B
|
|
chr11_-_41995688
|
29.510
|
|
Gabra1
|
gamma-aminobutyric acid (GABA) A receptor, subunit alpha 1
|
|
chr18_+_23573977
|
29.173
|
|
Dtna
|
dystrobrevin alpha
|
|
chr4_-_109959366
|
29.122
|
NM_001163399
NM_010488
|
Elavl4
|
ELAV (embryonic lethal, abnormal vision, Drosophila)-like 4 (Hu antigen D)
|
|
chr14_-_104213981
|
29.075
|
|
|
|